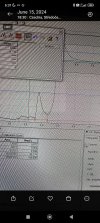
Here is the dimer peak clearly detectable in all 3 samples according to the European Pharmacopoeia method for Somatropin:
View attachment 286331
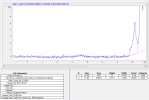
There is a large unknown peak (33% area @ 214nm) which appears after the monomer peak (visible on the chromatogram that we kindly provide as standard on all our reports). We are waiting on mass spec installation for proper verification -- however considering the large size, the ease at which it could be removed via purification, and its periodic presence in samples -- we believe it to be an excipient.
View attachment 286336
Based on the above absorbance plot, if area normalization were performed in the 240 - 250nm range, then the unknown would equate to ~3.5% area and the dimer peak would disappear. Unsure why janoshik does it this way, but other than the dimer and this unknown, there are no other peaks present.